Identifying common transcriptome signatures of cancer by interpreting deep learning models, Genome Biology
Por um escritor misterioso
Last updated 08 junho 2024
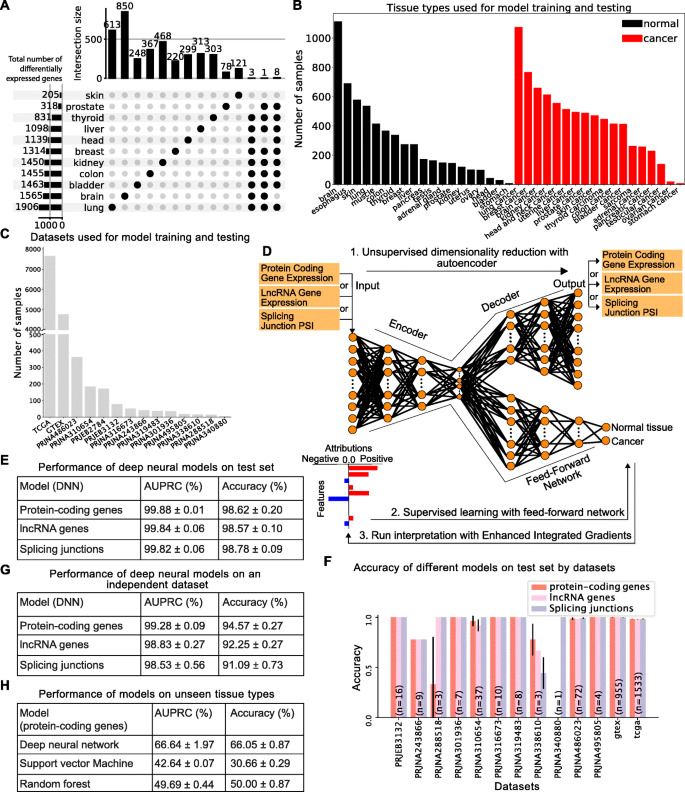
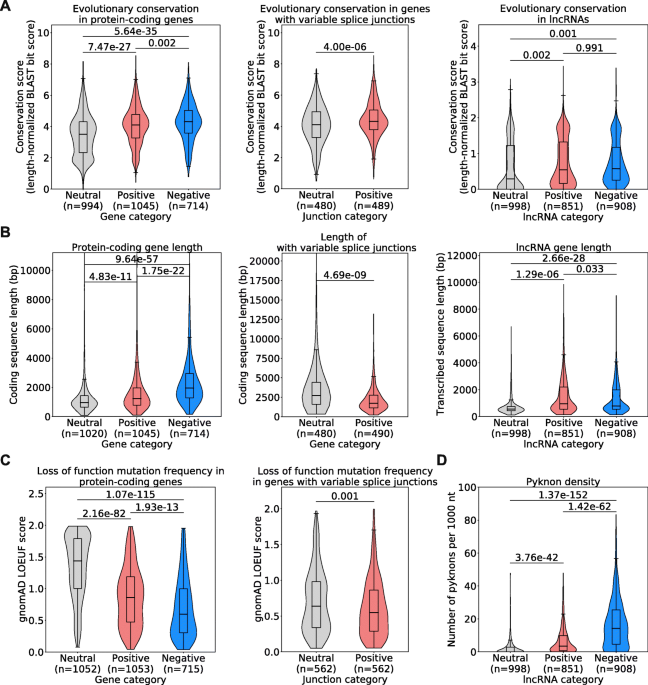
Background Cancer is a set of diseases characterized by unchecked cell proliferation and invasion of surrounding tissues. The many genes that have been genetically associated with cancer or shown to directly contribute to oncogenesis vary widely between tumor types, but common gene signatures that relate to core cancer pathways have also been identified. It is not clear, however, whether there exist additional sets of genes or transcriptomic features that are less well known in cancer biology but that are also commonly deregulated across several cancer types. Results Here, we agnostically identify transcriptomic features that are commonly shared between cancer types using 13,461 RNA-seq samples from 19 normal tissue types and 18 solid tumor types to train three feed-forward neural networks, based either on protein-coding gene expression, lncRNA expression, or splice junction use, to distinguish between normal and tumor samples. All three models recognize transcriptome signatures that are consistent across tumors. Analysis of attribution values extracted from our models reveals that genes that are commonly altered in cancer by expression or splicing variations are under strong evolutionary and selective constraints. Importantly, we find that genes composing our cancer transcriptome signatures are not frequently affected by mutations or genomic alterations and that their functions differ widely from the genes genetically associated with cancer. Conclusions Our results highlighted that deregulation of RNA-processing genes and aberrant splicing are pervasive features on which core cancer pathways might converge across a large array of solid tumor types.
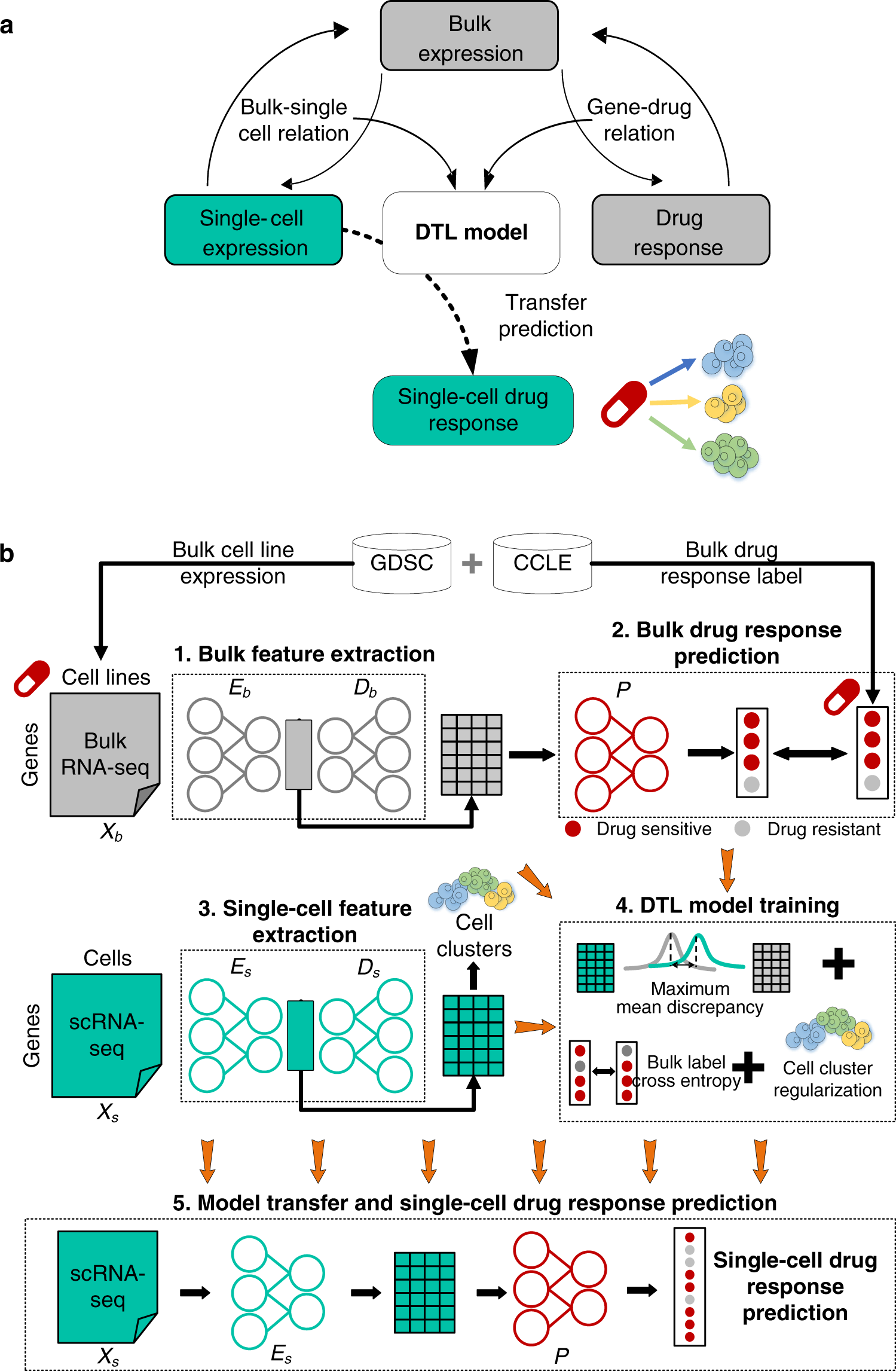
Deep transfer learning of cancer drug responses by integrating

Splicing signature database development to delineate cancer

Cancers, Free Full-Text

Identifying common transcriptome signatures of cancer by

Identifying functional regulatory mutation blocks by integrating
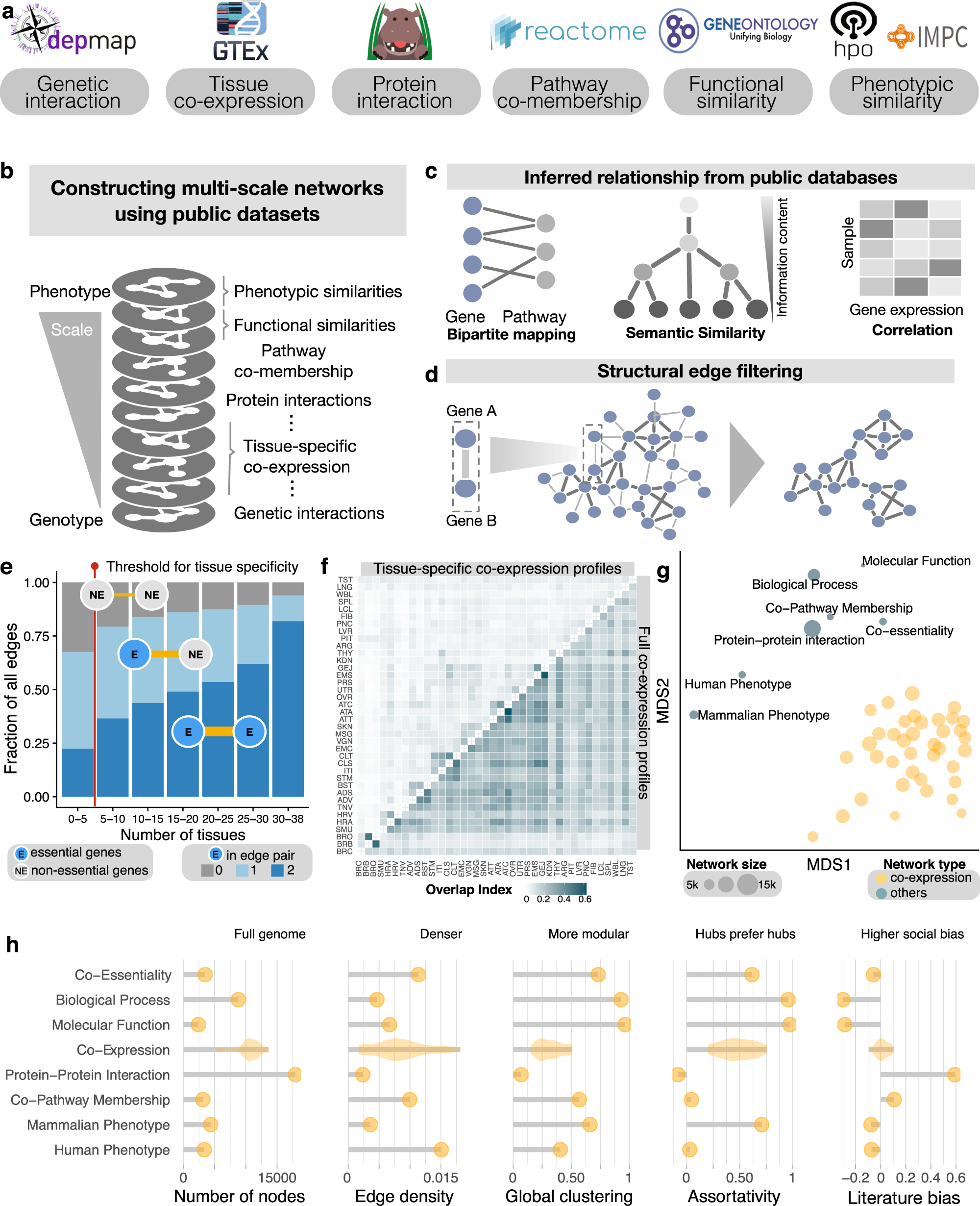
Network analysis reveals rare disease signatures across multiple
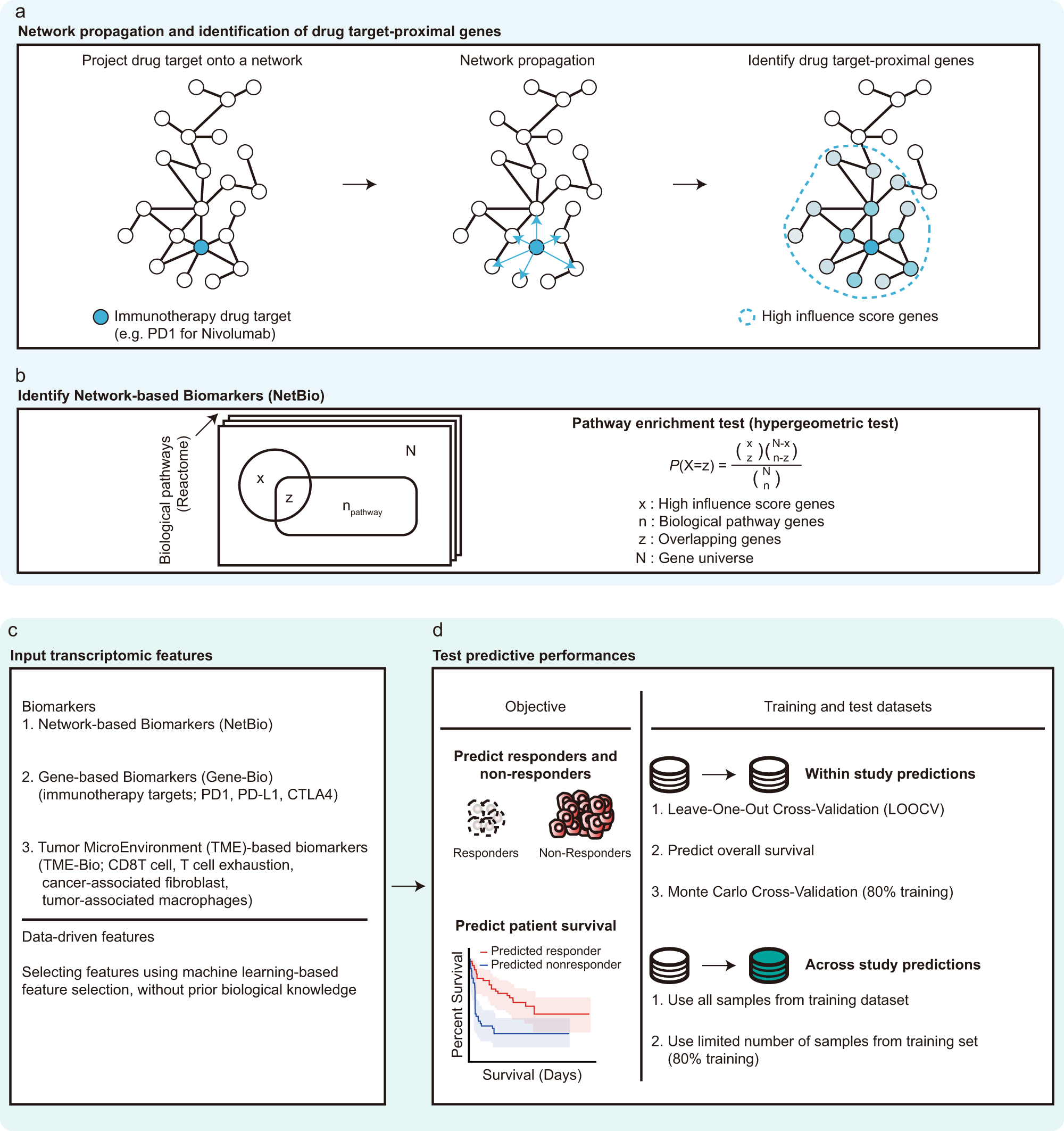
Network-based machine learning approach to predict immunotherapy

Biology, Free Full-Text
Identifying common transcriptome signatures of cancer by

The biological interpretation of deep neural network approach
Cancers, Free Full-Text
PDF) Identifying common transcriptome signatures of cancer by
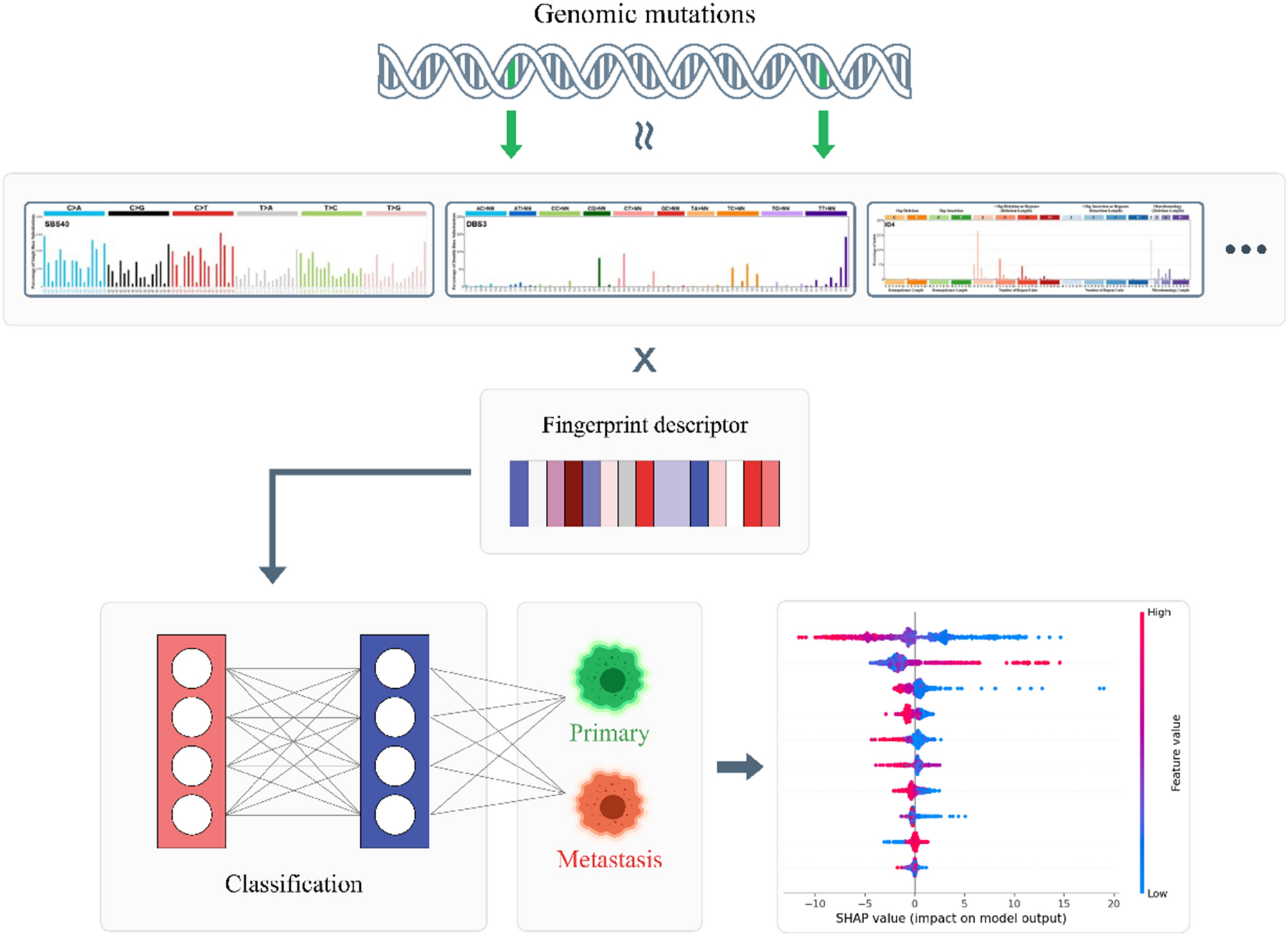
Deep learning model accurately classifies metastatic tumors from
Recomendado para você
-
 Test your brain power on Zoom in August! - General News - News08 junho 2024
Test your brain power on Zoom in August! - General News - News08 junho 2024 -
![PDF] Low-Dimensional Structure in the Space of Language Representations is Reflected in Brain Responses](https://d3i71xaburhd42.cloudfront.net/89711a7e91ad81874a5a0b1c3359bb67b27c0578/15-Table1-1.png) PDF] Low-Dimensional Structure in the Space of Language Representations is Reflected in Brain Responses08 junho 2024
PDF] Low-Dimensional Structure in the Space of Language Representations is Reflected in Brain Responses08 junho 2024 -
 Recent advancements in understanding fin regeneration in zebrafish - Sehring - 2020 - WIREs Developmental Biology - Wiley Online Library08 junho 2024
Recent advancements in understanding fin regeneration in zebrafish - Sehring - 2020 - WIREs Developmental Biology - Wiley Online Library08 junho 2024 -
 AI is helping scientists explain our brain08 junho 2024
AI is helping scientists explain our brain08 junho 2024 -
 Cabin Fever Word Search Puzzle – General Store of Minnetonka08 junho 2024
Cabin Fever Word Search Puzzle – General Store of Minnetonka08 junho 2024 -
 The 2021 Hyundai Santa Fe - Mid-Sized Favorite with Safety on the Brain08 junho 2024
The 2021 Hyundai Santa Fe - Mid-Sized Favorite with Safety on the Brain08 junho 2024 -
 Saatnya mencari cuan! (Brain Test Level 367) - CadeMedia08 junho 2024
Saatnya mencari cuan! (Brain Test Level 367) - CadeMedia08 junho 2024 -
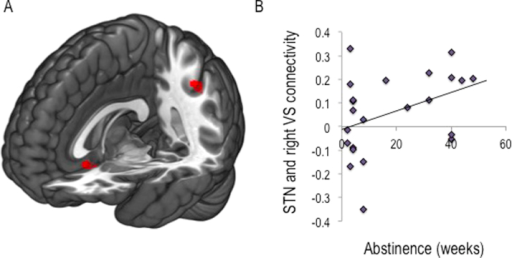 Subthalamic nucleus connectivity in binge drinkers and08 junho 2024
Subthalamic nucleus connectivity in binge drinkers and08 junho 2024 -
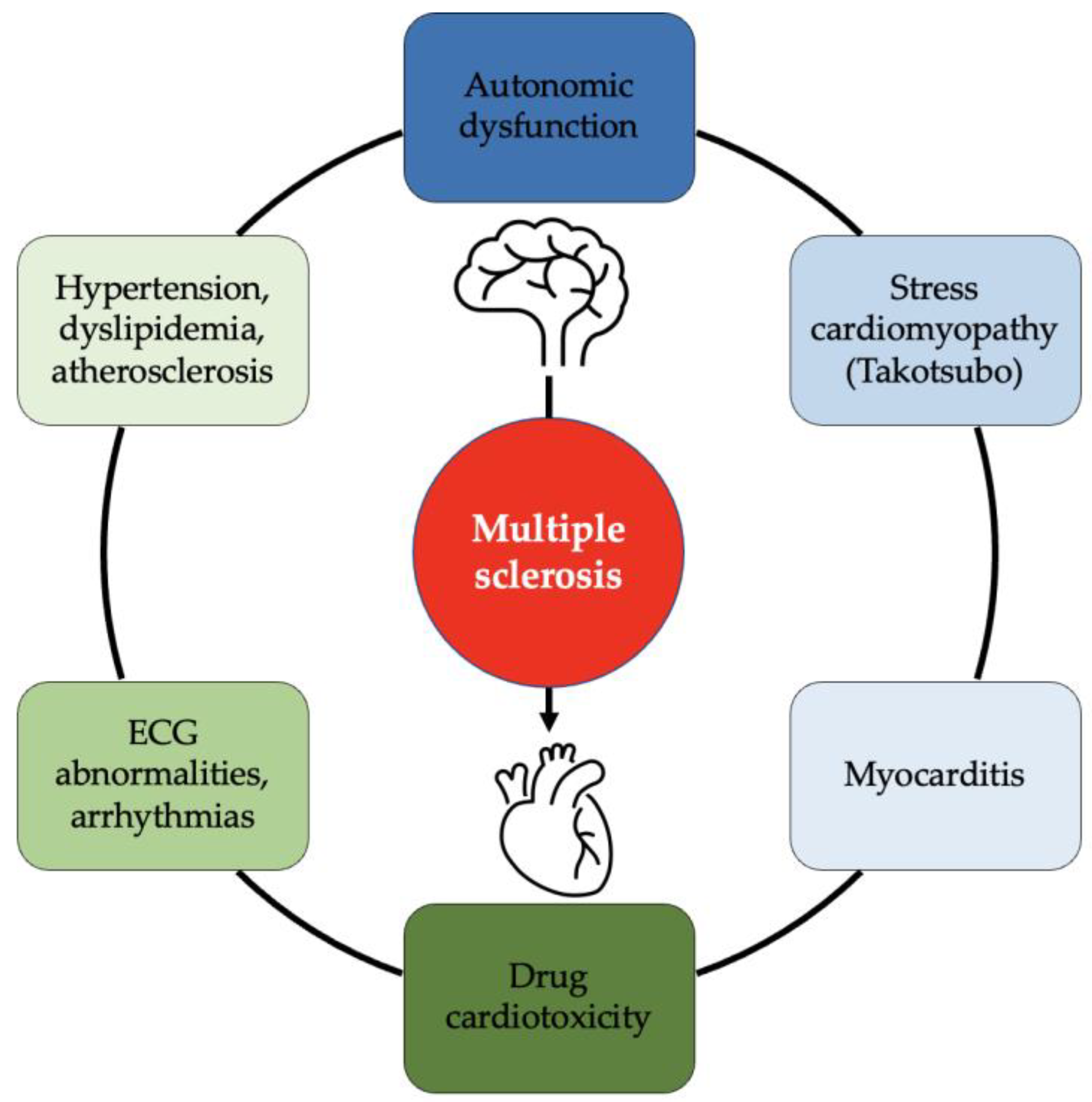 JCDD, Free Full-Text08 junho 2024
JCDD, Free Full-Text08 junho 2024 -
 Effectiveness of management strategies for uninvestigated dyspepsia: systematic review and network meta-analysis08 junho 2024
Effectiveness of management strategies for uninvestigated dyspepsia: systematic review and network meta-analysis08 junho 2024
você pode gostar
-
49% of 'Squid Game: The Challenge' fans want Player 287 (Mai) to08 junho 2024
-
Lista de Comandos MSC e CPL-Digite no Executar08 junho 2024
-
 I drew door 12 from popular roblox horror game: Doors : r/doors_roblox08 junho 2024
I drew door 12 from popular roblox horror game: Doors : r/doors_roblox08 junho 2024 -
 Buy Livewire + Whitesnake UK tickets, Livewire + Whitesnake UK tour details, Livewire + Whitesnake UK reviews08 junho 2024
Buy Livewire + Whitesnake UK tickets, Livewire + Whitesnake UK tour details, Livewire + Whitesnake UK reviews08 junho 2024 -
 Universal Range Box08 junho 2024
Universal Range Box08 junho 2024 -
 BAGAGEIRO DO GRAU SLIM TITAN 160 – Stunt Race Brasil08 junho 2024
BAGAGEIRO DO GRAU SLIM TITAN 160 – Stunt Race Brasil08 junho 2024 -
 Naruto Descobre que Boruto e Sarada se Tornaram um Casal após o Timeskip - Boruto Next Generation08 junho 2024
Naruto Descobre que Boruto e Sarada se Tornaram um Casal após o Timeskip - Boruto Next Generation08 junho 2024 -
Garten of Banban Pelúcia Jumbo Josh Opila presente de natal para crianças08 junho 2024
-
 Kimetsu no Yaiba: horario y dónde ver el episodio 6 de la Temporada 308 junho 2024
Kimetsu no Yaiba: horario y dónde ver el episodio 6 de la Temporada 308 junho 2024 -
 Pokemon Ash's Quest Fan Made Hack GBA Gameboy Advance, Canada08 junho 2024
Pokemon Ash's Quest Fan Made Hack GBA Gameboy Advance, Canada08 junho 2024

